Researchers across the world are working round the clock to find cures and a vaccine for the potent SARS-CoV-2 coronavirus which causes COVID-19. One of the biggest challenges that the researchers face is the prevention of the pathogen's reproduction. However, a new study by researchers from the University of Lübeck could aid in the development of drugs that can target the protein that regulates the virus' replication.
Through the study, the authors decoded the three-dimensional (3D) architecture of the key protease of the highly infectious disease using X-ray light from the BESSY II facility in Berlin. A concentrated examination of the protein's 3D architecture will enable the development of drugs that can impede the virus' reproduction.
Using BESSY II to decode the 3D architecture
A crucial protein is involved in the multiplication of the lethal virus — the viral main protease, which is also known as 3CLpro or Mpro. "An attractive drug target among coronaviruses is the main protease (Mpro, 3CLpro), due to its essential role in processing the polyproteins that are translated from the viral RNA," the authors wrote.
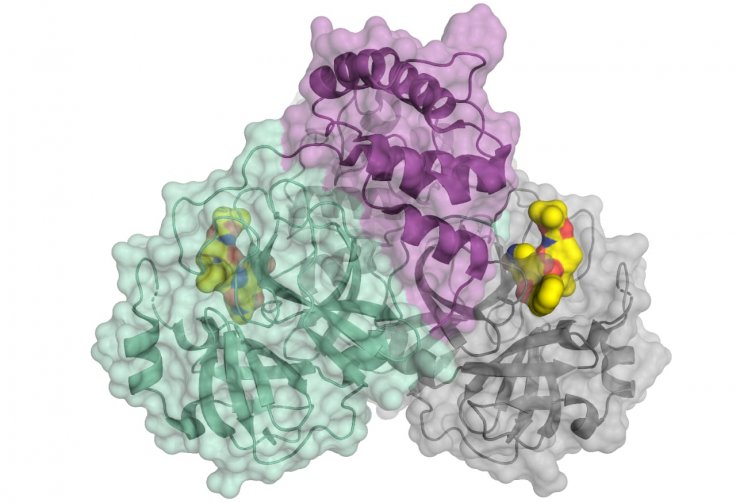
To decode the structure of the protein, the scientists used the high-intensity X-ray light from the BESSY II facility. BESSY stands for the 'Berlin Electron Storage Ring Society for Synchrotron Radiation'. BESSY II is a third generation synchrotron radiation facility — which is an advanced particle accelerator. It is operated by the Helmholtz-Zentrum Berlin (HZB).
How is the 3D structure studied?
Dr. Manfred Weiss, head of Research Group Macromolecular Crystallography (MX) at HZB, said: "For such issues of highest relevance, we can offer fast track access to our instruments." MX is a specialized technique using which various biological molecules such as viruses, nucleic acids and proteins at a highly advanced resolution. Using the facility's MX instrument, tiny crystals of proteins can be studied using extremely bright X-ray light.
In the images obtained, information regarding the 3D architecture of the protein molecules is contained. Using computer algorithms, its electron density and the complex structure of the protein molecule is calculated.
Aiding target-specific drugs
The most important application that the model can enable is the provision of a definitive starting point in the development of drugs that can curtail the reproductive mechanism of the virus. Using the 3D model, researchers can develop drugs that could target specific regions of the vital protein and disable the viruses' reproductive capacity.
So far 311,989 cases have been reported across 169 countries, with 13,407 casualties. Several concurrent research projects across the world are ongoing to find a remedy for the highly contagious disease.









